Despite the growing interest in Big Data and alternative data sources, household surveys remain the gold-standard for the production of official statistics. It should then come as no surprise that a diverse selection of packages have been developed to facilitate working with survey data in R. # Introduction Two of the most popular packages in this regard are sampling and survey for drawing survey samples and point/variance estimation, respectively, under complex sample designs. Yet, while both packages are part of the standard survey analyst’s toolbox, they are not the most user-friendly packages to work with (especially survey) but that is by no fault of their own. Both packages offer a bewildering array of options meant to support even the most arcane of survey techniques and it’s not easy to encapsulate the underlying complexity of all those methods behind a simple interface. Another issue with those packages is that they don’t fit neatly into tidy workflows though recent work on srvyr is trying to remedy that.
Our aim in this brief tutorial is to illustrate how the most common sample designs and estimation procedures can be carried out with little more than basic dplyr verbs. The sampling approach presented here was inspired by this post by Jennifer Bryan.
Let’s first start by constructing a synthetic census dataset to serve as our sampling frame.
library(tidyverse)
library(fabricatr)
set.seed(1)
refpop <-
fabricate(
ea = add_level(333),
hh =
add_level(
N = round(rnorm(length(ea), 100, 25)),
safety = c("Not safe", "Safe")[1+draw_binary_icc(clusters = ea, prob = .4, ICC = .1)],
pcexp = draw_normal_icc(clusters = ea, mean = 5000, sd = 1000, ICC = .1)))
hcpop <-
fabricate(
ea = add_level(667),
hh =
add_level(
N = round(rnorm(length(ea), 100, 25)),
safety = c("Not safe", "Safe")[1+draw_binary_icc(prob = .7, clusters = ea, ICC = .1)],
pcexp = draw_normal_icc(clusters = ea, mean = 10000, sd = 1500, ICC = .1)))
universe <-
bind_rows("Refugees" = refpop, "Host community" = hcpop, .id = "strata") %>%
as_tibble()That’s a population of about 100,000 households that is one-third refugees and two-thirds host community members (think of a small refugee camp and its neighboring villages). The population has been divided into enumeration areas with an average of 100 households each and we have two indicators on our synthetic population: safety, a binary indicator of whether the household feels safe or not in their neighborhood, and pcexp for an estimate of household per-capita expenditure. Both measures exhibit some degree of clustering as would be expected.
Here’s what the data looks like:
universe %>% head(10)## # A tibble: 10 x 5
## strata ea hh safety pcexp
## <chr> <chr> <chr> <chr> <dbl>
## 1 Refugees 001 00001 Safe 4447.
## 2 Refugees 001 00002 Not safe 4537.
## 3 Refugees 001 00003 Safe 3581.
## 4 Refugees 001 00004 Safe 5272.
## 5 Refugees 001 00005 Safe 5008.
## 6 Refugees 001 00006 Safe 7176.
## 7 Refugees 001 00007 Safe 6766.
## 8 Refugees 001 00008 Not safe 5269.
## 9 Refugees 001 00009 Safe 5631.
## 10 Refugees 001 00010 Safe 5308.Quick Aside: Nested Tables
What makes dplyr especially useful for working with survey samples is its support for nested tables. Table nesting comes in handy whenever you’re working with hierarchical data and want to run operations on entities at different levels of the hierarchy. The trick that allows this is that tables in R are just lists of equal-length items that store column values, but since lists can store any data type, including other lists, we can store entire tables in table cells.
The following is a perfectly valid data.frame in R:
(ex <-
tribble(
~parent, ~children,
"10s", tibble(x = 10:19),
"20s", tibble(x = 20:29),
"30s", tibble(x = 30:39)))## # A tibble: 3 x 2
## parent children
## <chr> <list>
## 1 10s <tibble [10 × 1]>
## 2 20s <tibble [10 × 1]>
## 3 30s <tibble [10 × 1]>And dplyr’s nest()/unnest() functions do all the hard-work of packing/unpacking data to allow us to move up and down the grouping hierarchy.
Unnesting moves us down:
(tmp <- ex %>% unnest(children))## # A tibble: 30 x 2
## parent x
## <chr> <int>
## 1 10s 10
## 2 10s 11
## 3 10s 12
## 4 10s 13
## 5 10s 14
## 6 10s 15
## 7 10s 16
## 8 10s 17
## 9 10s 18
## 10 10s 19
## # … with 20 more rowsWhile nesting takes us up:
tmp %>% group_nest(parent)## # A tibble: 3 x 2
## parent data
## <chr> <list<tbl_df[,1]>>
## 1 10s [10 × 1]
## 2 20s [10 × 1]
## 3 30s [10 × 1]It should be easy to see how this could help when working with nested populations.
Sampling
We’re now ready to dive into sampling.
Simple Random Sampling
Simple random sampling is the cornerstone foundation of all sample designs. It’s implemented in dplyr using sample_n(). Here’s a draw of 1000 households:
universe %>% sample_n(size = 1000)## # A tibble: 1,000 x 5
## strata ea hh safety pcexp
## <chr> <chr> <chr> <chr> <dbl>
## 1 Host community 128 12486 Safe 11196.
## 2 Host community 308 30522 Safe 5842.
## 3 Refugees 264 26485 Not safe 3529.
## 4 Host community 625 62127 Not safe 8179.
## 5 Host community 667 66311 Safe 9265.
## 6 Host community 259 25738 Safe 10715.
## 7 Host community 354 34970 Safe 11805.
## 8 Host community 172 16941 Safe 6745.
## 9 Host community 130 12736 Safe 9834.
## 10 Host community 476 47157 Safe 10941.
## # … with 990 more rowsStratified Sampling
On to a slightly more sophisticated sample design. The beauty of dplyr is that it allows you to express in code the same exact steps you would follow when describing the method:
universe %>%
# partition the population into strata
group_nest(strata) %>%
# for each stratum...
rowwise() %>%
mutate(
# calculate the required sample size
samplesz = round(1000*nrow(data)/nrow(universe)),
# then draw the sample
sample = list(sample_n(data, size = samplesz))) %>%
select(-c(data, samplesz)) %>%
# et voila!
unnest(sample)## # A tibble: 1,000 x 5
## strata ea hh safety pcexp
## <chr> <chr> <chr> <chr> <dbl>
## 1 Host community 628 62409 Not safe 12276.
## 2 Host community 110 10667 Not safe 7144.
## 3 Host community 455 44932 Safe 9399.
## 4 Host community 125 12189 Not safe 7934.
## 5 Host community 584 57910 Safe 9732.
## 6 Host community 301 29859 Safe 9707.
## 7 Host community 455 44944 Safe 6846.
## 8 Host community 309 30631 Not safe 9303.
## 9 Host community 100 09825 Safe 8350.
## 10 Host community 289 28624 Safe 7961.
## # … with 990 more rowsThat’s it for stratified sampling! For disproportionate stratification, just make sure to adjust samplesz accordingly.
Cluster Sampling
Clustering is fairly similar except this time we sample from the grouped table rather than within it.
universe %>%
# group the population by cluster
group_nest(ea) %>%
# draw the desired number of clusters
sample_n(size = 10) %>%
# and revert to a form that we can work with.
unnest(data)## # A tibble: 1,672 x 5
## ea strata hh safety pcexp
## <chr> <chr> <chr> <chr> <dbl>
## 1 634 Host community 62829 Not safe 9626.
## 2 634 Host community 62830 Not safe 8277.
## 3 634 Host community 62831 Not safe 12510.
## 4 634 Host community 62832 Not safe 11408.
## 5 634 Host community 62833 Not safe 11559.
## 6 634 Host community 62834 Not safe 9817.
## 7 634 Host community 62835 Safe 10844.
## 8 634 Host community 62836 Not safe 13357.
## 9 634 Host community 62837 Safe 10284.
## 10 634 Host community 62838 Not safe 13240.
## # … with 1,662 more rowsWe could have also passed any measure of size to sample_n()’s weight argument for unequal probability sampling.
Multi-stage Sampling
Armed with the basics, we can compose the pieces to construct arbitrarily complex designs as we wish.
Here, for example, is the familiar two-stage design with probability proportional to size selection at first stage and equal sample allocation across strata:
mssample <- function(universe) {
samplesz <- 1000
strata_cnt <- length(unique(universe$strata))
hh_per_cluster <- 10
clusters_per_stratum <- samplesz/hh_per_cluster/strata_cnt
msdata <-
universe %>%
group_nest(strata, ea) %>%
rowwise() %>%
mutate(psusz = nrow(data)) %>%
group_nest(strata) %>%
rowwise() %>%
mutate(stratasz = sum(data$psusz))
stage1 <-
msdata %>%
rowwise() %>%
mutate(sample = list(tibble(slice_sample(data, n = clusters_per_stratum, weight_by = data[[1]]$psusz)))) %>%
select(-data) %>%
unnest(sample)
stage2 <-
stage1 %>%
rowwise() %>%
mutate(sample = list(sample_n(data, size = hh_per_cluster))) %>%
select(-data) %>%
unnest(sample)
s <-
stage2 %>%
mutate(
p1 = psusz/stratasz, p2 = hh_per_cluster/psusz,
p = p1 * p2,
wt = 1/p, wt = wt/sum(wt) * length(wt)) %>%
select(-stratasz, -psusz, -p1, -p2, -p)
s
}
(s <- mssample(universe))## # A tibble: 1,000 x 6
## strata ea hh safety pcexp wt
## <chr> <chr> <chr> <chr> <dbl> <dbl>
## 1 Host community 039 03884 Not safe 10981. 1.33
## 2 Host community 039 03852 Not safe 7743. 1.33
## 3 Host community 039 03909 Not safe 9167. 1.33
## 4 Host community 039 03886 Safe 10630. 1.33
## 5 Host community 039 03811 Safe 12105. 1.33
## 6 Host community 039 03802 Not safe 9659. 1.33
## 7 Host community 039 03923 Not safe 9134. 1.33
## 8 Host community 039 03794 Not safe 11369. 1.33
## 9 Host community 039 03835 Safe 8881. 1.33
## 10 Host community 039 03915 Safe 10969. 1.33
## # … with 990 more rowsWe’ve stored this design and its output so we can reuse them in the second half of this tutorial. Also note that we’ve added normalized weights to our sample in the last step so that we could draw valid inferences from our sample.
Estimation
We’ll now shift our attention to estimation.
Point Estimation
Point estimation refers to the calculation of sums, averages, ratios, etc… from sample data.
For estimates involving categorical variables, dplyr’s count() already supports a wt argument for weighted calculations.
s %>% count(strata, safety, wt = wt) %>% group_by(strata) %>% mutate(p = n/sum(n)*100)## # A tibble: 4 x 4
## # Groups: strata [2]
## strata safety n p
## <chr> <chr> <dbl> <dbl>
## 1 Host community Not safe 186. 28.
## 2 Host community Safe 478. 72
## 3 Refugees Not safe 216. 64.2
## 4 Refugees Safe 121. 35.8Which is basically what we had specified when constructing our synthetic data: 70% of the host community feel safe in their neighborhoods whereas only 40% of the refugee population share the sentiment.
Actually, since our sample is self-weighting within each strata, the results would have come back the same with or without weights. Let’s look at the overall totals to see why weights are needed:
left_join(
s %>% count(safety, wt = wt, name = "n.weighted") %>% mutate(p.weighted = n.weighted/sum(n.weighted)*100),
s %>% count(safety, name = "n.unweighted") %>% mutate(p.unweighted = n.unweighted/sum(n.unweighted)*100))## # A tibble: 2 x 5
## safety n.weighted p.weighted n.unweighted p.unweighted
## <chr> <dbl> <dbl> <int> <dbl>
## 1 Not safe 402. 40.2 461 46.1
## 2 Safe 598. 59.8 539 53.9Now compare that to the census:
universe %>% count(safety) %>% mutate(p = n/sum(n)*100)## # A tibble: 2 x 3
## safety n p
## <chr> <int> <dbl>
## 1 Not safe 40673 40.7
## 2 Safe 59311 59.3And you could see that our weighted estimates are more in line with what we should have gotten.
What about point estimates involved continuous variables? Unfortunately there’s no convenience function similar to count() for those. That means we’ll have to fall back on the usual group+summarize workflow:
s %>% group_by(strata) %>% summarize(pcexp = weighted.mean(pcexp, wt))## # A tibble: 2 x 2
## strata pcexp
## <chr> <dbl>
## 1 Host community 9947.
## 2 Refugees 5025.Ground-truth:
universe %>% group_by(strata) %>% summarize(pcexp = mean(pcexp))## # A tibble: 2 x 2
## strata pcexp
## <chr> <dbl>
## 1 Host community 9977.
## 2 Refugees 4979.Close enough, with slight deviations because of sampling error… which brings us to the last part of this tutorial.
Variance Estimation
When producing estimates from sample data, we’re normally interested in more than just the point estimates. We also want to know how accurate our estimates are. This is the domain of variance estimation.
Let’s start by producing the actual sampling distribution of our mean per-capita expenditure statistic. We can do that since we’re working with simulated data. It wouldn’t otherwise be possible in the real world.
samplingdist <-
map_dfr(
1:500,
function(x) {
mssample(universe) %>%
{
bind_rows(
group_by(., strata) %>% summarize(pcexp = weighted.mean(pcexp, wt)),
group_by(., strata = "Everyone") %>% summarize(pcexp = weighted.mean(pcexp, wt)))
}
})Which tells us that our point estimates and standard errors should be:
(est_universe <- samplingdist %>% group_by(strata) %>% summarize(est = mean(pcexp), se = sd(pcexp)))## # A tibble: 3 x 3
## strata est se
## <chr> <dbl> <dbl>
## 1 Everyone 8295. 66.1
## 2 Host community 9979. 93.5
## 3 Refugees 4976. 60.0We had gotten to those point estimates in the previous section. Our job now is to try to reproduce the standard error se from nothing but the sample data.
Most software approaches variance estimation using analytical methods grounded in some smart mathematical approximations. We’ll take a more (brute-force) computational approach based on the bootstrap here. The bootstrap, for those not familiar with it, is the idea of resampling with replacement from sample data to approximate the sampling distribution of the target statistic. See section 2 of this article for more details on the how and why of bootstrapping.
We could implement the bootstrap ourselves using rerun() and sample_n() with replace = TRUE but instead we’re going to pull in tidymodels and use rsample::bootstraps(). It’s more convenient, idiomatic, and you’ll probably have tidymodels loaded in your workspace anyway if you’re planning on running any models on your survey data.
library(tidymodels)
b <- s %>% group_nest(strata, ea) %>% bootstraps(times = 500, strata = strata)
b <-
b %>%
rowwise() %>%
mutate(
stats =
analysis(splits) %>%
unnest(data) %>%
{
bind_rows(
group_by(., strata) %>% summarize(pcexp = weighted.mean(pcexp, wt)),
group_by(., strata = "Everyone") %>% summarize(pcexp = weighted.mean(pcexp, wt)))
} %>%
list()
)
(est_sample <-
b %>%
select(stats) %>%
unnest(stats) %>%
group_by(strata) %>%
summarize(est = mean(pcexp), se = sd(pcexp)))## # A tibble: 3 x 3
## strata est se
## <chr> <dbl> <dbl>
## 1 Everyone 8290. 59.7
## 2 Host community 9947. 85.1
## 3 Refugees 5024. 56.9The key thing to keep in mind is that the bootstrap sample should resemble the original sample in the way it is drawn. Meaning, if we had drawn clusters from within strata in the original sample, then we should resample clusters within strata for the bootstrap.
Let’s see how our estimates stack up against the actual values we had calculated earlier.
bind_rows("sampling distribution" = est_universe, "bootstrap" = est_sample, .id = "src") %>%
ggplot(aes(x = est, y = strata, xmin = est - 2*se, xmax = est + 2*se, color = src)) +
geom_point(position = position_dodge(-.5), size = 3) +
geom_errorbarh(position = position_dodge(-.5), height = .5) +
theme_minimal() +
labs(x = NULL, y = NULL, color = NULL,
title = "Estimated per-capita expenditures",
subtitle = "Mean & 95% confidence interval")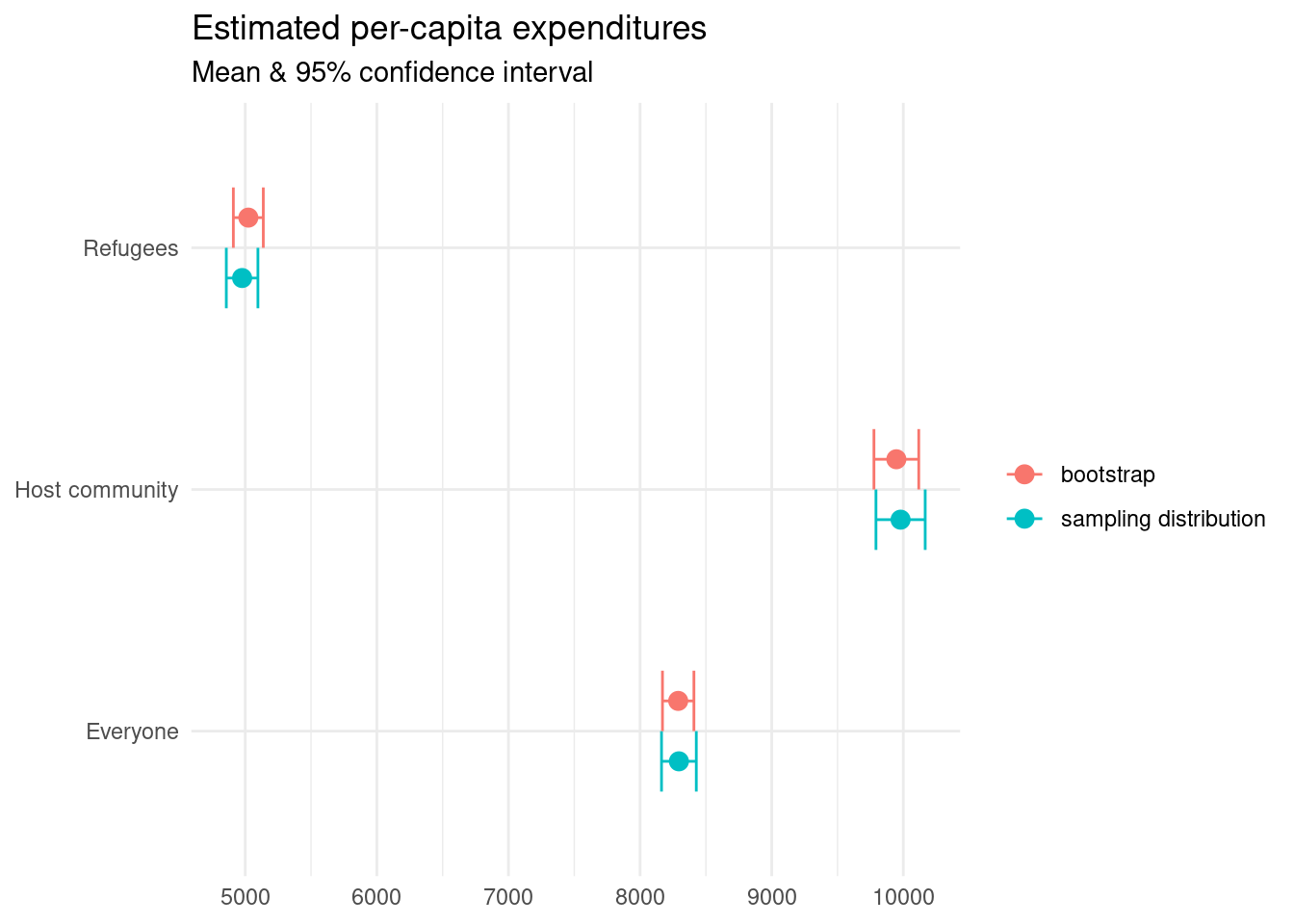
Confirming that our estimates line up with what they should be.
Share this post
Twitter
Google+
Facebook
Reddit
LinkedIn
StumbleUpon
Pinterest
Email